Am J Phys Anthropol DOI: 10.1002/ajpa.21591
Ecogeographical associations between climate and human body composition: Analyses based on anthropometry and skinfolds
Jonathan C.K. Wells et al.
Abstract
In the 19th century, two “ecogeographical rules” were proposed hypothesizing associations of climate with mammalian body size and proportions. Data on human body weight and relative leg length support these rules; however, it is unknown whether such associations are attributable to lean tissue (the heat-producing component) or fat (energy stores). Data on weight, height, and two skinfold thickness were obtained from the literature for 137 nonindustrialized populations, providing 145 male and 115 female individual samples. A variety of indices of adiposity and lean mass were analyzed. Preliminary analyses indicated secular increases in skinfolds in men but not women, and associations of age and height with lean mass in both sexes. Decreasing annual temperature was associated with increasing body mass index (BMI), and increasing triceps but not subscapular skinfold. After adjusting for skinfolds, decreasing temperature remained associated with increasing BMI. These results indicate that colder environments favor both greater peripheral energy stores, and greater lean mass. Contrasting results for triceps and subscapular skinfolds might be due to adaptive strategies either constraining central adiposity in cold environments to reduce cardiovascular risk, or favoring central adiposity in warmer environments to maintain energetic support of the immune system. Polynesian populations were analyzed separately and contradicted all of the climate trends, indicating support for the hypothesis that they are cold-adapted despite occupying a tropical region. It is unclear whether such associations emerge through natural selection or through trans-generational and life-course plasticity. These findings nevertheless aid understanding of the wide variability in human physique and adiposity.
Link
December 30, 2011
December 29, 2011
Chinese, Korean, Japanese (genetic edition)
My 2006 post on facial composites of Chinese, Korean, and Japanese women is, surprisingly, the most widely read single entry of this blog. People still occasionally guess "who is who" in that post, five years later.
As I was going through the list of the Dodecad populations, I realized that there are 5+ participants in each of the Korean, Japanese, and Chinese groups. So, it seemed like a simple exercise to see whether the relatively high success rate of people's guesses could be corroborated using the DNA data.
Below is the MDS plot; there are 9 Chinese, 5 Japanese, 5 Koreans in the Dodecad Project; I have also added 30 HapMap Chinese (CHB) and Japanese (JPT):
Only the first MDS dimension showed deviation from normality according to a Shapiro-Wilk test. Using MCLUST, that dimension was enough (as can be seen from the above figure) to infer the presence of 3 clusters which corresponded to the 3 groups, with 100% correct assignments.
Interestingly, when I did not use the extra HapMap individuals, MCLUST did not split Koreans from Chinese. This goes to show that the absence of apparent structure does not imply absence of structure. The extra Chinese and Japanese individuals helped flesh out the existing structure in these East Asian groups.
Below is the list of the Dodecad populations that are below the 5-individual limit:
If you belong to one of the above groups (all 4 grandparents) and have tested with either 23andMe or Family Finder, you are especially invited to contact me at dodecad@gmail.com (but do not send data right away!), about possible inclusion in the project.
For example, in the most recent Clusters Galore analysis, there was a generic "Balkan" cluster. Does this imply that Balkan ethnic groups cannot be distinguished from each other, or that sample sizes are simply not yet sufficient to make manifest the existing structure?
As I was going through the list of the Dodecad populations, I realized that there are 5+ participants in each of the Korean, Japanese, and Chinese groups. So, it seemed like a simple exercise to see whether the relatively high success rate of people's guesses could be corroborated using the DNA data.
Below is the MDS plot; there are 9 Chinese, 5 Japanese, 5 Koreans in the Dodecad Project; I have also added 30 HapMap Chinese (CHB) and Japanese (JPT):
Only the first MDS dimension showed deviation from normality according to a Shapiro-Wilk test. Using MCLUST, that dimension was enough (as can be seen from the above figure) to infer the presence of 3 clusters which corresponded to the 3 groups, with 100% correct assignments.
Interestingly, when I did not use the extra HapMap individuals, MCLUST did not split Koreans from Chinese. This goes to show that the absence of apparent structure does not imply absence of structure. The extra Chinese and Japanese individuals helped flesh out the existing structure in these East Asian groups.
Below is the list of the Dodecad populations that are below the 5-individual limit:
| Algerian_D | 4 | East_African_Various_D | 3 | Greek_Italian_D | 2 | Belgian_D | 1 | |||
| North_African_Jews_D | 4 | Danish_D | 3 | Swiss_German_D | 2 | Latvian_D | 1 | |||
| Slovenian_D | 4 | Tunisian_D | 3 | Szekler_D | 2 | Estonian_D | 1 | |||
| Mixed_Scandinavian_D | 4 | Austrian_D | 3 | Mandaean_D | 2 | Bangladesh_D | 1 | |||
| Moroccan_D | 4 | Saudi_D | 3 | Azeri_D | 2 | Yemenese_D | 1 | |||
| Serb_D | 4 | Pakistani_D | 3 | Czech_D | 2 | Sri_Lanka_D | 1 | |||
| Tatar_Various_D | 3 | Georgian_D | 2 | Hungarian_D | 1 | |||||
| Palestinian_D | 3 | Kazakh_D | 2 | Basque_D | 1 | |||||
| Romanian_D | 3 | Udmurt_D | 1 | |||||||
| Ukrainian_D | 1 | |||||||||
| Egyptian_D | 1 |
If you belong to one of the above groups (all 4 grandparents) and have tested with either 23andMe or Family Finder, you are especially invited to contact me at dodecad@gmail.com (but do not send data right away!), about possible inclusion in the project.
For example, in the most recent Clusters Galore analysis, there was a generic "Balkan" cluster. Does this imply that Balkan ethnic groups cannot be distinguished from each other, or that sample sizes are simply not yet sufficient to make manifest the existing structure?
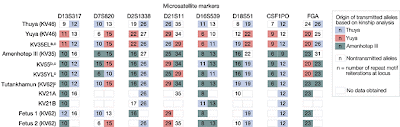
Forensic analysis of King Tut and his relatives
DNA Tribes has released an analysis, based on 8 forensic autosomal STR markers, of the "Amarna Pharaohs". The analysis is based on data from Ancestry and Pathology in King Tutankhamun's Family.
The results of the DNA Tribes analysis can be seen below:
They seem to indicate that there is something definitely "African" about this collection of mummies. I have previously used PopAffiliator and STRUCTURE with CODIS markers. The results of that analysis suggest that even this small number of markers is sufficient to place a sample in a continental group with high accuracy, but insufficient to estimate levels of admixture. There is a new version of PopAffiliator, which, unfortunately, does not allow for incomplete data entry, and hence cannot be used to verify the results of the DNA Tribes analysis.
The DNA Tribes results are interesting, but may hinge upon a few marker values that are more prevalent in Africa than in Eurasia. Also, it is not clear which population(s) make up the "North African" group. It would be interesting to extract full genome sequences from Egyptian mummies in order to properly place them in the global genetic landscape.
Pictorial evidence in Egyptian art, as well as the statements of classical Greco-Roman authors strongly suggest that the ancient Egyptians occupied an intermediate position in the phenotypic continuum between Near Eastern and "Ethiopian" people. It is also clear that there was variation within ancient Egypt itself: geographic, temporal, and even perhaps social aspects of this variation may have existed. But these qualitative observations are no substitute for the harder type of evidence that can be provided by authentic ancient DNA.
Hopefully, the debate on the genetic identity of the ancient Egyptians can proceed on the basis of new data, although I am not holding my breath that this will happen anytime soon, both because of the fluid state of politics in Egypt itself, the existence of various fringe theories outside of Egypt, and, the rather controversial state of mummy DNA analysis itself.
They seem to indicate that there is something definitely "African" about this collection of mummies. I have previously used PopAffiliator and STRUCTURE with CODIS markers. The results of that analysis suggest that even this small number of markers is sufficient to place a sample in a continental group with high accuracy, but insufficient to estimate levels of admixture. There is a new version of PopAffiliator, which, unfortunately, does not allow for incomplete data entry, and hence cannot be used to verify the results of the DNA Tribes analysis.
The DNA Tribes results are interesting, but may hinge upon a few marker values that are more prevalent in Africa than in Eurasia. Also, it is not clear which population(s) make up the "North African" group. It would be interesting to extract full genome sequences from Egyptian mummies in order to properly place them in the global genetic landscape.
Pictorial evidence in Egyptian art, as well as the statements of classical Greco-Roman authors strongly suggest that the ancient Egyptians occupied an intermediate position in the phenotypic continuum between Near Eastern and "Ethiopian" people. It is also clear that there was variation within ancient Egypt itself: geographic, temporal, and even perhaps social aspects of this variation may have existed. But these qualitative observations are no substitute for the harder type of evidence that can be provided by authentic ancient DNA.
Hopefully, the debate on the genetic identity of the ancient Egyptians can proceed on the basis of new data, although I am not holding my breath that this will happen anytime soon, both because of the fluid state of politics in Egypt itself, the existence of various fringe theories outside of Egypt, and, the rather controversial state of mummy DNA analysis itself.
December 28, 2011
Genetic structure in China
After my experiment on Spain, I decided to carry out a similar experiment in China, for which there is a large number of regional/ethnic sub-populations.15 clusters were inferred with 22 MDS dimensions.
The Uygur are the clear outlier population, doubtlessly due to their substantial Caucasoid admixture and geographical position in Central Asia, a region that was traditionally at the outskirts of Chinese civilization. Other Altaic speakers (both Mongolic and Tungusic) are also divergent, as are the Dai/Lahu people from the China/Thailand/Laos area.
Interestingly, the Tujia people from Central China seem to be the ones most like the Han overall, with Hmongic Miaozu/She more like the southern Han.
The Uygur are the clear outlier population, doubtlessly due to their substantial Caucasoid admixture and geographical position in Central Asia, a region that was traditionally at the outskirts of Chinese civilization. Other Altaic speakers (both Mongolic and Tungusic) are also divergent, as are the Dai/Lahu people from the China/Thailand/Laos area.
Interestingly, the Tujia people from Central China seem to be the ones most like the Han overall, with Hmongic Miaozu/She more like the southern Han.
Craniofacial Differences Between Modern and Archaeological Asian Skeletal Populations (Ph.D. thesis)
From p. 169:
Craniofacial Differences Between Modern and Archaeological Asian Skeletal Populations
Chan, Wing Nam Joyce
The principal objective of this study is to perform a biological distance analysis of two Asian ethnic groups to better understand environmental factors influencing cranial shape and size. Cranial shape and size are influenced by both epigenetic and genetic factors, resulting in differences in crania over time. Cranial measurements can be used as a proxy for genetic data and to understand epigenetic factors affecting crania. Therefore, craniometrics can be used to determine differences between populations.
Ancient and modern Chinese and Thai skeletal populations were used for this biological distance analysis. The ancient Chinese population is from northern China at Anyang dating to the Shang Dynasty (1600BC-1046BC) while its modern counterpart is located in Hong Kong dating from 1977-1983. Individuals from both populations are thought to have belonged to the Han ethnic group and are possibly biologically related. Both Thai populations are located in northeastern Thailand, known as the Isaan region. The ancient Thai population from the Ban Chiang site is dated through the Pre-metal to Iron Age periods (2000 B.C.- 200 A.D.) while the modern population dates from 1970s to present. Data were collected on crania at 29 anthropologically accepted measurements to explore epigenetic and biological relationships between modern and ancient populations. Data were subjected to multiple multivariate statistical tests to understand causative agents for change and differences between populations.
These results suggest that modern and ancient Thai and Chinese populations have markedly different crania, especially in shape. However, correlated factors could not be identified in this study, primarily due to lack of historical data. Geographical, temporal, and climate variables such as temperature were tested against measures of biological distance with little to no correlation discovered. Interestingly, modern and ancient Chinese populations displayed the closest biological affinity, possibly due to similar environments and lack of genetic changes. Ban Chiang individuals were the most biologically distant from other populations, indicating possible genetic differences not yet understood. These genetic differences could indicate either that Ban Chiang individuals are not recently ancestral to the modern Thai population or a mass migration movement into northeast Thailand had occurred.
These results are interpreted to indicate that environmental factors have played a large role in altering cranial shape in these two ethnically Asian populations since genetic alteration in the areas has not been documented. Environmental factors have caused isometric changes in cranial shape as crania have become distinct from their ancestral counterparts. Cultural changes, such as diet shifts and modernization, are possible causative agents for these changes witnessed in these populations.
The findings of this study contribute to our understanding of human cranial variation for these two Asian groups, and to broader discussions of epigenetic and genetic relationships in the expression of cranial morphology. This research also contributes to the discussions of how biological distance in the crania has been influenced by epigenetic factors and ultimately how the peopling of modern Asia occurred.
Link
Since physical changes have been documented in these Asian populations, principal components analyses were conducted for certain variable groupings in each population to understand how crania shape were related. Based on visual inspection, ancient crania appears to be long and narrower compared to their more modern counterparts, and these tests will illustrate which portions of the crania are changing over time.p. 181:
Since physical differences were shown to be present between the two ethnic groups, shape and size changes were calculated to understand how the crania have changed over time. Principal components analysis was calculated using transformed log variables of the dataset to determine which measurements were changing. All populations appeared to be extremely divergent from one another, resulting in unequal variancecovariance matrices. Modern Chinese individuals appear to have a shorter head than their ancient counterpart while modern Thais tend to have narrower faces and heads than their ancestral group.p. 187:
None of the variable groupings show a close biological relationship between modern populations. However, the modern Thai population did show a closer biological relationship with the modern Chinese than with either archaeological population. One possible reason for this outcome is due to the influx of Chinese immigrants to Thailand that eventually became part of the gene pool. However, gene flow did not work in reverse with Thai people moving into China. For the most part Thai people remained ethnically Thai and Chinese people remained Chinese.p. 197:
Modern Chinese individuals appear to have narrower and longer crania, which is opposite than expected. Ancient Chinese crania have higher cranial vaults than their modern counterparts, which contributes to their divergence. Facial variables that appear to be most divergent are ones that measure for prognathism and upper facial height. Modern Chinese individuals have longer upper facial height but are not as prognathic as their ancient counterparts.
Craniofacial Differences Between Modern and Archaeological Asian Skeletal Populations
Chan, Wing Nam Joyce
The principal objective of this study is to perform a biological distance analysis of two Asian ethnic groups to better understand environmental factors influencing cranial shape and size. Cranial shape and size are influenced by both epigenetic and genetic factors, resulting in differences in crania over time. Cranial measurements can be used as a proxy for genetic data and to understand epigenetic factors affecting crania. Therefore, craniometrics can be used to determine differences between populations.
Ancient and modern Chinese and Thai skeletal populations were used for this biological distance analysis. The ancient Chinese population is from northern China at Anyang dating to the Shang Dynasty (1600BC-1046BC) while its modern counterpart is located in Hong Kong dating from 1977-1983. Individuals from both populations are thought to have belonged to the Han ethnic group and are possibly biologically related. Both Thai populations are located in northeastern Thailand, known as the Isaan region. The ancient Thai population from the Ban Chiang site is dated through the Pre-metal to Iron Age periods (2000 B.C.- 200 A.D.) while the modern population dates from 1970s to present. Data were collected on crania at 29 anthropologically accepted measurements to explore epigenetic and biological relationships between modern and ancient populations. Data were subjected to multiple multivariate statistical tests to understand causative agents for change and differences between populations.
These results suggest that modern and ancient Thai and Chinese populations have markedly different crania, especially in shape. However, correlated factors could not be identified in this study, primarily due to lack of historical data. Geographical, temporal, and climate variables such as temperature were tested against measures of biological distance with little to no correlation discovered. Interestingly, modern and ancient Chinese populations displayed the closest biological affinity, possibly due to similar environments and lack of genetic changes. Ban Chiang individuals were the most biologically distant from other populations, indicating possible genetic differences not yet understood. These genetic differences could indicate either that Ban Chiang individuals are not recently ancestral to the modern Thai population or a mass migration movement into northeast Thailand had occurred.
These results are interpreted to indicate that environmental factors have played a large role in altering cranial shape in these two ethnically Asian populations since genetic alteration in the areas has not been documented. Environmental factors have caused isometric changes in cranial shape as crania have become distinct from their ancestral counterparts. Cultural changes, such as diet shifts and modernization, are possible causative agents for these changes witnessed in these populations.
The findings of this study contribute to our understanding of human cranial variation for these two Asian groups, and to broader discussions of epigenetic and genetic relationships in the expression of cranial morphology. This research also contributes to the discussions of how biological distance in the crania has been influenced by epigenetic factors and ultimately how the peopling of modern Asia occurred.
Link
Southeast Asian origin of dogs (again)
PLoS ONE 6(12): e28496. doi:10.1371/journal.pone.0028496
Phylogenetic Distinctiveness of Middle Eastern and Southeast Asian Village Dog Y Chromosomes Illuminates Dog Origins
Sarah K. Brown et al.
Modern genetic samples are commonly used to trace dog origins, which entails untested assumptions that village dogs reflect indigenous ancestry or that breed origins can be reliably traced to particular regions. We used high-resolution Y chromosome markers (SNP and STR) and mitochondrial DNA to analyze 495 village dogs/dingoes from the Middle East and Southeast Asia, along with 138 dogs from >35 modern breeds to 1) assess genetic divergence between Middle Eastern and Southeast Asian village dogs and their phylogenetic affinities to Australian dingoes and gray wolves (Canis lupus) and 2) compare the genetic affinities of modern breeds to regional indigenous village dog populations. The Y chromosome markers indicated that village dogs in the two regions corresponded to reciprocally monophyletic clades, reflecting several to many thousand years divergence, predating the Neolithic ages, and indicating long-indigenous roots to those regions. As expected, breeds of the Middle East and East Asia clustered within the respective regional village dog clade. Australian dingoes also clustered in the Southeast Asian clade. However, the European and American breeds clustered almost entirely within the Southeast Asian clade, even sharing many haplotypes, suggesting a substantial and recent influence of East Asian dogs in the creation of European breeds. Comparison to 818 published breed dog Y STR haplotypes confirmed this conclusion and indicated that some African breeds reflect another distinct patrilineal origin. The lower-resolution mtDNA marker consistently supported Y-chromosome results. Both marker types confirmed previous findings of higher genetic diversity in dogs from Southeast Asia than the Middle East. Our findings demonstrate the importance of village dogs as windows into the past and provide a reference against which ancient DNA can be used to further elucidate origins and spread of the domestic dog.
Link
Phylogenetic Distinctiveness of Middle Eastern and Southeast Asian Village Dog Y Chromosomes Illuminates Dog Origins
Sarah K. Brown et al.
Modern genetic samples are commonly used to trace dog origins, which entails untested assumptions that village dogs reflect indigenous ancestry or that breed origins can be reliably traced to particular regions. We used high-resolution Y chromosome markers (SNP and STR) and mitochondrial DNA to analyze 495 village dogs/dingoes from the Middle East and Southeast Asia, along with 138 dogs from >35 modern breeds to 1) assess genetic divergence between Middle Eastern and Southeast Asian village dogs and their phylogenetic affinities to Australian dingoes and gray wolves (Canis lupus) and 2) compare the genetic affinities of modern breeds to regional indigenous village dog populations. The Y chromosome markers indicated that village dogs in the two regions corresponded to reciprocally monophyletic clades, reflecting several to many thousand years divergence, predating the Neolithic ages, and indicating long-indigenous roots to those regions. As expected, breeds of the Middle East and East Asia clustered within the respective regional village dog clade. Australian dingoes also clustered in the Southeast Asian clade. However, the European and American breeds clustered almost entirely within the Southeast Asian clade, even sharing many haplotypes, suggesting a substantial and recent influence of East Asian dogs in the creation of European breeds. Comparison to 818 published breed dog Y STR haplotypes confirmed this conclusion and indicated that some African breeds reflect another distinct patrilineal origin. The lower-resolution mtDNA marker consistently supported Y-chromosome results. Both marker types confirmed previous findings of higher genetic diversity in dogs from Southeast Asia than the Middle East. Our findings demonstrate the importance of village dogs as windows into the past and provide a reference against which ancient DNA can be used to further elucidate origins and spread of the domestic dog.
Link
The function of the Aterian
From the paper:
Shape Variation in Aterian Tanged Tools and the Origins of Projectile Technology: A Morphometric Perspective on Stone Tool Function
Radu Iovita
Abstract
Background Recent findings suggest that the North African Middle Stone Age technocomplex known as the Aterian is both much older than previously assumed, and certainly associated with fossils exhibiting anatomically modern human morphology and behavior. The Aterian is defined by the presence of ‘tanged’ or ‘stemmed’ tools, which have been widely assumed to be among the earliest projectile weapon tips. The present study systematically investigates morphological variation in a large sample of Aterian tools to test the hypothesis that these tools were hafted and/or used as projectile weapons.
Methodology/Principal Findings Both classical morphometrics and Elliptical Fourier Analysis of tool outlines are used to show that the shape variation in the sample exhibits size-dependent patterns consistent with a reduction of the tools from the tip down, with the tang remaining intact. Additionally, the process of reduction led to increasing side-to-side asymmetries as the tools got smaller. Finally, a comparison of shape-change trajectories between Aterian tools and Late Paleolithic arrowheads from the North German site of Stellmoor reveal significant differences in terms of the amount and location of the variation.
Conclusions/Significance The patterns of size-dependent shape variation strongly support the functional hypothesis of Aterian tools as hafted knives or scrapers with alternating active edges, rather than as weapon tips. Nevertheless, the same morphological patterns are interpreted as one of the earliest evidences for a hafting modification, and for the successful combination of different raw materials (haft and stone tip) into one implement, in itself an important achievement in the evolution of hominin technologies.
Link
The ability of human hunters to ‘kill at a distance’ [1], [2] is often considered one of the hallmarks of modern human behavior. Such an ability embodies the cultural transcendence of the human body's condition with the aid of technology and has deep implications for the self-understanding of our species's uniqueness in the animal kingdom. For this reason, the search for evidence of projectile weapon technologies in the Stone Age has superseded the search for evidence of mere hunting activities, the latter having slid in the background of pre-human hominin behavioral repertoire [3]–[5]. Because ‘safe hunting’ is considered to have given anatomically-modern humans a competitive advantage against Neandertals during the last Out-of-Africa event (e.g., [6], [7]), it is extremely important to rigorously examine claims for the existence of such technologies, even when the superficial examination of the morphology of a particular tool suggests a clear functional determination. Such is the case of the Aterian tanged (or stemmed) point, a type of stone tool found throughout North Africa in a variety of ecological, geographical, and chronological contexts within the African Middle Stone Age (MSA), and which exhibits a simple form that is sometimes reminiscent of stemmed arrowheads or spear points from much later time periods (Figure 1). ... The importance of correctly interpreting the function of Aterian stemmed tools is underlined by recent dating results, which suggests that, contrary to early assumptions, it could date to as early as MIS 5 and before [29]. More specifically, new dates from a series of sites, such Mugharet el-Aliya [30], Rhafas [31], Ifri n'Ammar [29], Dar-es-Soltan [32], and Contrebandiers [33] have demonstrated that tanged tools can be found in the earliest part of the North African Middle Stone Age, making them potentially the earliest evidence of prehistoric stone-tipped weaponry. However, the precise way in which they actually fit within a prehistoric technological system, including whether or not they were part of flying projectile armatures or thrusting spears, has never been rigorously determined, despite the crucial role that both projectiles and hafting are thought to play in the evolution of human cultural adaptations.PLoS ONE 6(12): e29029. doi:10.1371/journal.pone.0029029
...
Several lines of evidence point in the direction of progressive resharpening of Aterian tools in the same manner as edge-tools such as scrapers and cutting-tools. This does not per se rule out an initial use for some Aterian pieces as weapon tips, because the ultimate use of each individual tool must be determined by the examination of use-traces, and because each episode of retouch likely wipes out previous uses of the tool. However, the data presented here make a strong case for the claim that, in general, these tools were probably hafted and used repeatedly for tasks that resulted in the need to rejuvenate edges rather than point-tips. The comparison between excavated, mostly cave contexts, and surface sites reveals that they both contain similar reduction trajectories and shape variabilities of tanged tools. This indicates that the functional emphasis on the tools was similar during their use-life in the landscape and at the repeated-occupation sites, which contradicts the expectations of breakage and repair patterns associated with a use as projectile tips [62], [63].
...
It is thus possible that hafting was practiced on both sides of the Mediterranean Sea, but in different ways. Although some of the differences in technological innovation between archaic and modern humans that we observe at the continental and species level may be due to cognitive differences or to demographic factors influencing the spread and accumulation of information [79], [80], we must not forget the essentially functional character of toolkits. Especially when comparing and evaluating technologies at very large scales, functional responses to specific technological problems (such as prey size and behavior [1] or increased risk associated with prey frequency and ease of hunting (e.g., [81]–[84])) may trump other factors. Even if the ultimate cause underpinning technological change is a large-scale environmental phenomenon, such as a rapid cooling event, or the aridity of a newly-colonized area, we can understand these associated changes only by unraveling the constraints imposed on toolkits by the subjects of the actions for which the tools themselves were used. Thus, perhaps the better question to answer regarding the Aterian might not be if it represents the earliest hunting weapons technology, but rather, in what way it arose out of new challenges posed by the environments that characterized North Africa since MIS 5, and how it adapted during the almost 100 thousand years of occupation of this region.
Shape Variation in Aterian Tanged Tools and the Origins of Projectile Technology: A Morphometric Perspective on Stone Tool Function
Radu Iovita
Abstract
Background Recent findings suggest that the North African Middle Stone Age technocomplex known as the Aterian is both much older than previously assumed, and certainly associated with fossils exhibiting anatomically modern human morphology and behavior. The Aterian is defined by the presence of ‘tanged’ or ‘stemmed’ tools, which have been widely assumed to be among the earliest projectile weapon tips. The present study systematically investigates morphological variation in a large sample of Aterian tools to test the hypothesis that these tools were hafted and/or used as projectile weapons.
Methodology/Principal Findings Both classical morphometrics and Elliptical Fourier Analysis of tool outlines are used to show that the shape variation in the sample exhibits size-dependent patterns consistent with a reduction of the tools from the tip down, with the tang remaining intact. Additionally, the process of reduction led to increasing side-to-side asymmetries as the tools got smaller. Finally, a comparison of shape-change trajectories between Aterian tools and Late Paleolithic arrowheads from the North German site of Stellmoor reveal significant differences in terms of the amount and location of the variation.
Conclusions/Significance The patterns of size-dependent shape variation strongly support the functional hypothesis of Aterian tools as hafted knives or scrapers with alternating active edges, rather than as weapon tips. Nevertheless, the same morphological patterns are interpreted as one of the earliest evidences for a hafting modification, and for the successful combination of different raw materials (haft and stone tip) into one implement, in itself an important achievement in the evolution of hominin technologies.
Link
December 27, 2011
Y-chromosome of Emperor Cao Cao: O2
Thanks to the wonders of population genetics, not only do we have a good inference of the Y-haplogroups of Cao Cao and Cao Shen, but we can also falsify (?) the claim of descent of a Chinese emperor from 18-19 centuries ago.
That's 4x deeper in time than the Y-haplogroup of Nurhaci. It would be great if a similar study could produce conclusive results for Confucius descendants.
And, I'm sure that the burial sites of many important European historical figures are well-known, as are the lines of descent of many descendants of the old nobility. In a roundabout way, we obtained the Y-DNA of Louis XVI, and Tsar Nicholas II, but there is plenty more to be discovered.
J Hum Genet. 2011 Dec 22. doi: 10.1038/jhg.2011.147. [Epub ahead of print]
Present Y chromosomes reveal the ancestry of Emperor CAO Cao of 1800 years ago.
Wang C, Yan S, Hou Z, Fu W, Xiong M, Han S, Jin L, Li H.
Abstract Emperor CAO Cao (155AD-220AD) is one of the most famous persons in Chinese history that had changed the history of East Asia. He claimed to be a descendant of Marquis CAO Can and therefore was of aristocratic ancestry. However, this claim has been suspected for around 1800 years. Here, we collected some present clans with full records of 70-100 generations claimed to be descendants of CAO Cao or CAO Can, and validated them by comparing their Y chromosomes. Haplotype O2-M268 is the only one that is enriched significantly in the Emperor's claimed descendant clans (P=9.323 × 10(-5), odds ratio=12.72) and, therefore, is most likely to be that of the Emperor. Moreover, our analysis showed that the Y chromosome haplotype of the Emperor is different from that of the Marquis (Haplotype O3-002611). Therefore, Emperor CAO Cao's claim was not supported by genetic evidence. This study offers a successful showcase of the utility of genetics in studying the ancient history.
That's 4x deeper in time than the Y-haplogroup of Nurhaci. It would be great if a similar study could produce conclusive results for Confucius descendants.
And, I'm sure that the burial sites of many important European historical figures are well-known, as are the lines of descent of many descendants of the old nobility. In a roundabout way, we obtained the Y-DNA of Louis XVI, and Tsar Nicholas II, but there is plenty more to be discovered.
J Hum Genet. 2011 Dec 22. doi: 10.1038/jhg.2011.147. [Epub ahead of print]
Present Y chromosomes reveal the ancestry of Emperor CAO Cao of 1800 years ago.
Wang C, Yan S, Hou Z, Fu W, Xiong M, Han S, Jin L, Li H.
Abstract Emperor CAO Cao (155AD-220AD) is one of the most famous persons in Chinese history that had changed the history of East Asia. He claimed to be a descendant of Marquis CAO Can and therefore was of aristocratic ancestry. However, this claim has been suspected for around 1800 years. Here, we collected some present clans with full records of 70-100 generations claimed to be descendants of CAO Cao or CAO Can, and validated them by comparing their Y chromosomes. Haplotype O2-M268 is the only one that is enriched significantly in the Emperor's claimed descendant clans (P=9.323 × 10(-5), odds ratio=12.72) and, therefore, is most likely to be that of the Emperor. Moreover, our analysis showed that the Y chromosome haplotype of the Emperor is different from that of the Marquis (Haplotype O3-002611). Therefore, Emperor CAO Cao's claim was not supported by genetic evidence. This study offers a successful showcase of the utility of genetics in studying the ancient history.
Stature of prehistoric Europeans
J Hum Evol. 2011 Dec 22. [Epub ahead of print]
Stature estimation from complete long bones in the Middle Pleistocene humans from the Sima de los Huesos, Sierra de Atapuerca (Spain).
Carretero JM, Rodríguez L, García-González R, Arsuaga JL, Gómez-Olivencia A, Lorenzo C, Bonmatí A, Gracia A, Martínez I, Quam R.
Abstract Systematic excavations at the site of the Sima de los Huesos (SH) in the Sierra de Atapuerca (Burgos, Spain) have allowed us to reconstruct 27 complete long bones of the human species Homo heidelbergensis. The SH sample is used here, together with a sample of 39 complete Homo neanderthalensis long bones and 17 complete early Homo sapiens (Skhul/Qafzeh) long bones, to compare the stature of these three different human species. Stature is estimated for each bone using race- and sex-independent regression formulae, yielding an average stature for each bone within each taxon. The mean length of each long bone from SH is significantly greater (p < 0.05) than the corresponding mean values in the Neandertal sample. The stature has been calculated for male and female specimens separately, averaging both means to calculate a general mean. This general mean stature for the entire sample of long bones is 163.6 cm for the SH hominins, 160.6 cm for Neandertals and 177.4 cm for early modern humans. Despite some overlap in the ranges of variation, all mean values in the SH sample (whether considering isolated bones, the upper or lower limb, males or females or more complete individuals) are larger than those of Neandertals. Given the strong relationship between long bone length and stature, we conclude that SH hominins represent a slightly taller population or species than the Neandertals. However, compared with living European Mediterranean populations, neither the Sima de los Huesos hominins nor the Neandertals should be considered 'short' people. In fact, the average stature within the genus Homo seems to have changed little over the course of the last two million years, since the appearance of Homo ergaster in East Africa. It is only with the emergence of H. sapiens, whose earliest representatives were 'very tall', that a significant increase in stature can be documented.
Link
Stature estimation from complete long bones in the Middle Pleistocene humans from the Sima de los Huesos, Sierra de Atapuerca (Spain).
Carretero JM, Rodríguez L, García-González R, Arsuaga JL, Gómez-Olivencia A, Lorenzo C, Bonmatí A, Gracia A, Martínez I, Quam R.
Abstract Systematic excavations at the site of the Sima de los Huesos (SH) in the Sierra de Atapuerca (Burgos, Spain) have allowed us to reconstruct 27 complete long bones of the human species Homo heidelbergensis. The SH sample is used here, together with a sample of 39 complete Homo neanderthalensis long bones and 17 complete early Homo sapiens (Skhul/Qafzeh) long bones, to compare the stature of these three different human species. Stature is estimated for each bone using race- and sex-independent regression formulae, yielding an average stature for each bone within each taxon. The mean length of each long bone from SH is significantly greater (p < 0.05) than the corresponding mean values in the Neandertal sample. The stature has been calculated for male and female specimens separately, averaging both means to calculate a general mean. This general mean stature for the entire sample of long bones is 163.6 cm for the SH hominins, 160.6 cm for Neandertals and 177.4 cm for early modern humans. Despite some overlap in the ranges of variation, all mean values in the SH sample (whether considering isolated bones, the upper or lower limb, males or females or more complete individuals) are larger than those of Neandertals. Given the strong relationship between long bone length and stature, we conclude that SH hominins represent a slightly taller population or species than the Neandertals. However, compared with living European Mediterranean populations, neither the Sima de los Huesos hominins nor the Neandertals should be considered 'short' people. In fact, the average stature within the genus Homo seems to have changed little over the course of the last two million years, since the appearance of Homo ergaster in East Africa. It is only with the emergence of H. sapiens, whose earliest representatives were 'very tall', that a significant increase in stature can be documented.
Link
Lack of significant population structure in Spain
I took the Iberian Spanish (IBS) regional populations, and ran multidimensional scaling on them (left). Most of the populations form a tight cluster, with Basques and Canary Islanders having averages further removed from the main cluster.
It should be noted that the Canarias sample consists of only two individuals (big red dots): one removed from the main cluster, one in the midst of individuals from Castilla Y Leon (small red dots).
MCLUST analysis using 1 dimension, reveals 2 clusters: one consisting of Pais Vasco individuals (blue dots), the other of everyone else, with one Aragonese and one Cantabrian individual showing mixed probabilities between the two clusters.
The overall impression is that there may be additional population structure here (e.g., for Canarians or Galicians), but the sample sizes are not sufficient to make additional clusters unambiguously evident, except in the case of Basques vs. non-Basques.
It should be noted that the Canarias sample consists of only two individuals (big red dots): one removed from the main cluster, one in the midst of individuals from Castilla Y Leon (small red dots).
MCLUST analysis using 1 dimension, reveals 2 clusters: one consisting of Pais Vasco individuals (blue dots), the other of everyone else, with one Aragonese and one Cantabrian individual showing mixed probabilities between the two clusters.
The overall impression is that there may be additional population structure here (e.g., for Canarians or Galicians), but the sample sizes are not sufficient to make additional clusters unambiguously evident, except in the case of Basques vs. non-Basques.
December 25, 2011
Merry Christmas
Ἡ Παρθένος σήμερον, τὸν ὑπερούσιον τίκτει,
καὶ ἡ γῆ τὸ Σπήλαιον, τῷ ἀπροσίτῳ προσάγει.
Ἄγγελοι μετὰ Ποιμένων δοξολογοῦσι.
Μάγοι δὲ μετὰ ἀστέρος ὁδοιποροῦσι.
δι' ἡμᾶς γὰρ ἐγεννήθη, Παιδίον νέον, ὁ πρὸ αἰώνων Θεός.
καὶ ἡ γῆ τὸ Σπήλαιον, τῷ ἀπροσίτῳ προσάγει.
Ἄγγελοι μετὰ Ποιμένων δοξολογοῦσι.
Μάγοι δὲ μετὰ ἀστέρος ὁδοιποροῦσι.
δι' ἡμᾶς γὰρ ἐγεννήθη, Παιδίον νέον, ὁ πρὸ αἰώνων Θεός.
December 23, 2011
Multiple origins of Russian mtDNA
The genetic distances from the Russians to the Europeanlanguage groups indicate that the gene pool of present-day Russians bears the influence of Slavic, Baltic,Finno-Ugric and, to a lesser extent, Germanic groups, aswell as Iranian and Turkic groups....
The results of this study strongly suggest that the impact of the pre-Slavic (Finno-Ugric) population on the East European Plain is the most important factor for the northward and southward differentiation of the present-day Russian gene pool. This explanation supports the view proposing the genetic influence of Finno-Ugrians on the formation of the northern regions of Russia, which was inferred from mtDNA marker studies of some Russian populations (Grzybowski et al., 2007) and Y-chromosome analysis (Balanovsky et al., 2008).
Being quite distant from the Finno-Ugric group, the Southern Russians consequently differ from the Northern Russians in their closeness to the Germanic group. This difference indicates that the Germanic people played a significant role in the development of the southern, but not the northern segment of the Russian gene pool. In general, the Germanic influence on the formation of the Russians is not as obvious as the impact of the Slavic, Baltic, and Finno-Ugric people. However, strong interactions between the Germanic and Slavic tribes have been found in archeological materials dating from the mid-first millennium B.C. to the early first millennium A.D. These interactions were the strongest on the northern coast of the Black Sea, in the area of the multiethnic Chernyakhov archeological culture (second to fifth centuries A.D.). In the second half of the first millennium A.D., the descendants of this culture colonized the southern regions of the historical Russian area (Sedov, 1994, 1995). However, there is no evidence in the historical literature of the interaction between the Germanic tribes and the Slavs (and later, the Russians) after the Slavic colonization of the East European Plain. Therefore, the Germanic influence could not have occurred after the early part of the first millennium A.D., which was before the eastward Slavic migration (Sedov, 1994, 1995). Apparently, the impact of the Germanic people on the Chernyakhov Slavs affected the gene pool of modern Southern Russians, consequently differentiating them from the Northern Russians (Fig. 6).Am J Phys Anthropol DOI: 10.1002/ajpa.21649
Russian ethnic history inferred from mitochondrial DNA diversity
Irina Morozova et al.
With the aim of gaining insight into the genetic history of the Russians, we have studied mitochondrial DNA diversity among a number of modern Russian populations. Polymorphisms in mtDNA markers (HVS-I and restriction sites of the coding region) of populations from 14 regions within present-day European Russia were investigated. Based on analysis of the mitochondrial gene pool geographic structure, we have identified three different elements in it and a vast “intermediate” zone between them. The analysis of the genetic distances from these elements to the European ethnic groups revealed the main causes of the Russian mitochondrial gene pool differentiation. The investigation of this pattern in historic perspective showed that the structure of the mitochondrial gene pool of the present-day Russians largely conforms to the tribal structure of the medieval Slavs who laid the foundation of modern Russians. Our results indicate that the formation of the genetic diversity currently observed among Russians can be traced to the second half of the first millennium A.D., the time of the colonization of the East European Plain by the Slavic tribes. Patterns of diversity are explained by both the impact of the native population of the East European Plain and by genetic differences among the early Slavs.
Link
Cranial nonmetric traits of Garamantes
Am J Phys Anthropol DOI: 10.1002/ajpa.21645
Sahara: Barrier or corridor? Nonmetric cranial traits and biological affinities of North African late holocene populations
Efthymia Nikita et al.
The Garamantes flourished in southwestern Libya, in the core of the Sahara Desert ∼3,000 years ago and largely controlled trans-Saharan trade. Their biological affinities to other North African populations, including the Egyptian, Algerian, Tunisian and Sudanese, roughly contemporary to them, are examined by means of cranial nonmetric traits using the Mean Measure of Divergence and Mahalanobis D2 distance. The aim is to shed light on the extent to which the Sahara Desert inhibited extensive population movements and gene flow. Our results show that the Garamantes possess distant affinities to their neighbors. This relationship may be due to the Central Sahara forming a barrier among groups, despite the archaeological evidence for extended networks of contact. The role of the Sahara as a barrier is further corroborated by the significant correlation between the Mahalanobis D2 distance and geographic distance between the Garamantes and the other populations under study. In contrast, no clear pattern was observed when all North African populations were examined, indicating that there was no uniform gene flow in the region.
Link
Sahara: Barrier or corridor? Nonmetric cranial traits and biological affinities of North African late holocene populations
Efthymia Nikita et al.
The Garamantes flourished in southwestern Libya, in the core of the Sahara Desert ∼3,000 years ago and largely controlled trans-Saharan trade. Their biological affinities to other North African populations, including the Egyptian, Algerian, Tunisian and Sudanese, roughly contemporary to them, are examined by means of cranial nonmetric traits using the Mean Measure of Divergence and Mahalanobis D2 distance. The aim is to shed light on the extent to which the Sahara Desert inhibited extensive population movements and gene flow. Our results show that the Garamantes possess distant affinities to their neighbors. This relationship may be due to the Central Sahara forming a barrier among groups, despite the archaeological evidence for extended networks of contact. The role of the Sahara as a barrier is further corroborated by the significant correlation between the Mahalanobis D2 distance and geographic distance between the Garamantes and the other populations under study. In contrast, no clear pattern was observed when all North African populations were examined, indicating that there was no uniform gene flow in the region.
Link
December 22, 2011
Voice pitch and semen quality
The effect was small, but, nonetheless quite interesting. If semen quality is linked to the probability of a pregnancy per copulation, and if voice attractiveness is linked to the expected number of copulations, then it's easy to see how a tradeoff between voice attractiveness and semen quality might work.
PLoS ONE 6(12): e29271. doi:10.1371/journal.pone.0029271
Low Pitched Voices Are Perceived as Masculine and Attractive but Do They Predict Semen Quality in Men?
Leigh W. Simmons et al.
Women find masculinity in men's faces, bodies, and voices attractive, and women's preferences for men's masculine features are thought to be biological adaptations for finding a high quality mate. Fertility is an important aspect of mate quality. Here we test the phenotype-linked fertility hypothesis, which proposes that male secondary sexual characters are positively related to semen quality, allowing females to obtain direct benefits from mate choice. Specifically, we examined women's preferences for men's voice pitch, and its relationship with men's semen quality. Consistent with previous voice research, women judged lower pitched voices as more masculine and more attractive. However men with lower pitched voices did not have better semen quality. On the contrary, men whose voices were rated as more attractive tended to have lower concentrations of sperm in their ejaculate. These data are more consistent with a trade off between sperm production and male investment in competing for and attracting females, than with the phenotype-linked fertility hypothesis.
Link
PLoS ONE 6(12): e29271. doi:10.1371/journal.pone.0029271
Low Pitched Voices Are Perceived as Masculine and Attractive but Do They Predict Semen Quality in Men?
Leigh W. Simmons et al.
Women find masculinity in men's faces, bodies, and voices attractive, and women's preferences for men's masculine features are thought to be biological adaptations for finding a high quality mate. Fertility is an important aspect of mate quality. Here we test the phenotype-linked fertility hypothesis, which proposes that male secondary sexual characters are positively related to semen quality, allowing females to obtain direct benefits from mate choice. Specifically, we examined women's preferences for men's voice pitch, and its relationship with men's semen quality. Consistent with previous voice research, women judged lower pitched voices as more masculine and more attractive. However men with lower pitched voices did not have better semen quality. On the contrary, men whose voices were rated as more attractive tended to have lower concentrations of sperm in their ejaculate. These data are more consistent with a trade off between sperm production and male investment in competing for and attracting females, than with the phenotype-linked fertility hypothesis.
Link
December 20, 2011
Non-destructive extraction of ancient nuclear and mitochondrial DNA
Ancient DNA often involves pulverizing a tooth or other bone sample. This is undesirable, since the material is often irreplaceable, and there are competing interests: e.g., the gain of the highly risky and possibly non-conclusive extraction of DNA from a tooth vs. the loss of this tooth for research by dental anthropologists.
Apart from the details of the new methodology, the paper also contains new ancient mtDNA and autosomal data:
Am J Phys Anthropol DOI: 10.1002/ajpa.21647
Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA
Deborah A. Bolnick et al.
Museum curators and living communities are sometimes reluctant to permit ancient DNA (aDNA) studies of human skeletal remains because the extraction of aDNA usually requires the destruction of at least some skeletal material. Whether these views stem from a desire to conserve precious materials or an objection to destroying ancestral remains, they limit the potential of aDNA research. To help address concerns about destructive analysis and to minimize damage to valuable specimens, we describe a nondestructive method for extracting DNA from ancient human remains. This method can be used with both teeth and bone, but it preserves the structural integrity of teeth much more effectively than that of bone. Using this method, we demonstrate that it is possible to extract both mitochondrial and nuclear DNA from human remains dating between 300 BC and 1600 AD. Importantly, the method does not expose the remains to hazardous chemicals, allowing them to be safely returned to curators, custodians, and/or owners of the samples. We successfully amplified mitochondrial DNA from 90% of the individuals tested, and we were able to analyze 1–9 nuclear loci in 70% of individuals. We also show that repeated nondestructive extractions from the same tooth can yield amplifiable mitochondrial and nuclear DNA. The high success rate of this method and its ability to yield DNA from samples spanning a wide geographic and temporal range without destroying the structural integrity of the sampled material may make possible the genetic study of skeletal collections that are not available for destructive analysis.
Link
Apart from the details of the new methodology, the paper also contains new ancient mtDNA and autosomal data:
Am J Phys Anthropol DOI: 10.1002/ajpa.21647
Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA
Deborah A. Bolnick et al.
Museum curators and living communities are sometimes reluctant to permit ancient DNA (aDNA) studies of human skeletal remains because the extraction of aDNA usually requires the destruction of at least some skeletal material. Whether these views stem from a desire to conserve precious materials or an objection to destroying ancestral remains, they limit the potential of aDNA research. To help address concerns about destructive analysis and to minimize damage to valuable specimens, we describe a nondestructive method for extracting DNA from ancient human remains. This method can be used with both teeth and bone, but it preserves the structural integrity of teeth much more effectively than that of bone. Using this method, we demonstrate that it is possible to extract both mitochondrial and nuclear DNA from human remains dating between 300 BC and 1600 AD. Importantly, the method does not expose the remains to hazardous chemicals, allowing them to be safely returned to curators, custodians, and/or owners of the samples. We successfully amplified mitochondrial DNA from 90% of the individuals tested, and we were able to analyze 1–9 nuclear loci in 70% of individuals. We also show that repeated nondestructive extractions from the same tooth can yield amplifiable mitochondrial and nuclear DNA. The high success rate of this method and its ability to yield DNA from samples spanning a wide geographic and temporal range without destroying the structural integrity of the sampled material may make possible the genetic study of skeletal collections that are not available for destructive analysis.
Link
2012 looking good for my predictions
I wrote:
All indications point in the direction that the Afrasian/Palaeoafrican theory is about to be confirmed. I purposefully decided to name the major recent component in our species' ancestry "Afrasian", because I did not want to take a strong stand on where this component originated (Africa or Asia). My reticence to jump on the recent Out-of-Africa bandwagon with both feet, seems to have been well-justified, as Out-of-Arabia seems to be an increasingly strong possibility: from the LiveScience piece, once again:
Many mysteries about human origins will be solved thanks to the advent of full genome sequencing. Hammer et al. found archaic admixture in Africans on just 61 genomic regions, each about ~20kb in length.From LiveScience via Razib:
I'm willing to bet that once scientists turn their attentions to full genomes, they will have substantial and indisputable evidence for genetic divergence between stretches of human DNA that simply too deep to be explained in a conventional Out-of-Africa timeframe.
If there was substantial archaic admixture in Africa c. 35ka, according to Hammer et al.'s estimate, and coinciding with the (intrusive?) appearance of Upper Paleolithic modern humans such as Hofmeyr, then full genome sequencing will provide the smoking gun evidence for it. Such an event would simultaneously solve many mysteries about the African population, such as its apparent higher effective population size, greater allele diversity, and recombination rate.
Our species might have also hybridized with a now-extinct lineage of humanity before leaving Africa, according to findings this year from Hammer and his colleagues. Approximately 2 percent of contemporary African DNA might have come from a lineage that first diverged from the ancestors of modern humans about 700,000 years ago. For context, the Neanderthal lineage diverged from ours within the past 500,000 years, while the first signs of anatomically modern human features emerged only about 200,000 years ago.
Hammer noted that he and his colleagues were very conservative with their analysis, only looking for lineages that diverged even more from modern humans than Neanderthals. "It's possible there may be others we can detect that are more closely related to modern humans," Hammer told LiveScience.
"We've probably just scratched the surface of what we might find," Hammer added. "We only looked at a small number of regions of the genome. This coming year, you'll see a lot of progress made with full genome data. This year, we should be able to confirm what we found and go way beyond that."I started talking about "Afrasians" mixing it up with "Palaeoafricans" as a major cause for African genetic diversity back in 2005. From a 2006 treatment:
It is clear that the small early modern human population must have inhabited a correspondingly small geographical region, so it is not surprising that in their movements within Africa they would have interbred with the pre-existing humans. After all, humans lived in Africa for a long time before the emergence of the moderns, and there is no reason to believe that all the African branches of humanity were wiped out to be replaced by the advancing moderns.
I predict that in the coming years, we will learn much more about the different strata of genetic ancestry contained in Africans, as well as Europeans and East Asians. Note, also, that there is no candidate for the source population of the archaic contribution of West Africans. This, again, is not surprising, because western Africa has a much less advantageous climate than eastern Africa for bone preservation, in addition to being less well researched. Even in Europe, where anthropological science is the oldest, and cave surveys have been numerous, there are still only a handful of well-preserved Neanderthal specimens. Hopefully, some of the archaics of Africa remain to be discovered.Some of these archaics have indeed been found.
All indications point in the direction that the Afrasian/Palaeoafrican theory is about to be confirmed. I purposefully decided to name the major recent component in our species' ancestry "Afrasian", because I did not want to take a strong stand on where this component originated (Africa or Asia). My reticence to jump on the recent Out-of-Africa bandwagon with both feet, seems to have been well-justified, as Out-of-Arabia seems to be an increasingly strong possibility: from the LiveScience piece, once again:
"I hope that our findings will stimulate research in South Asia — India in particular — to find the remains of early anatomically modern humans in that part of the world,"archaeologist Hans-Peter Uerpmann from Eberhard Karls University in Tubingen, Germany, told LiveScience.
"Our focus this year will be on gathering evidence to reconstruct the paleoclimate in southern Arabia during the ice age that lasted between 75,000 and 60,000 years ago," paleolithic archaeologist Jeffrey Rose at the University of Birmingham in England told LiveScience. This will help researchers determine how friendly or hostile the climate was back then "to help understand the fate of these early humans on the Arabian Peninsula."
If these ancient peoples eventually died off in Arabia, they would just be a failed migration out of Africa. However, if they survived, they may be the ancestors "to all non-African people living on Earth," Rose said. "Only further exploration throughout Arabia will answer these questions."These Middle Stone Age inhabitants of Arabia may not just be the ancestors only of everyone outside Africa, but of many within Africa itself.
Syphilis spread from the Americas in the last 500 years
The discovery of the New World by Columbus and the ones that followed him launched an era of exchange between the two hemispheres that saw Old World populations and material culture transported west, and New World culture transported east. The arrival and spread of European diseases among Native Americans is well-documented, but there is at least one disease that spread in the opposite direction: syphilis. Previous research has suggested that syphilitic symptoms could be discerned in pre-Columbian populations of the Old World,
A new comprehensive survey of the evidence, however, suggests that syphilis did come from the Americas due to Columbus' voyage, and that the evidence advanced for a pre-Columbian presence of the disease is not solid.
More:
Yrbk Phys Anthropol 54:99–133, 2011.
The origin and antiquity of syphilis revisited: An Appraisal of Old World pre-Columbian evidence for treponemal infection
Kristin N. Harper et al.
For nearly 500 years, scholars have argued about the origin and antiquity of syphilis. Did Columbus bring the disease from the New World to the Old World? Or did syphilis exist in the Old World before 1493? Here, we evaluate all 54 published reports of pre-Columbian, Old World treponemal disease using a standardized, systematic approach. The certainty of diagnosis and dating of each case is considered, and novel information pertinent to the dating of these cases, including radiocarbon dates, is presented. Among the reports, we did not find a single case of Old World treponemal disease that has both a certain diagnosis and a secure pre-Columbian date. We also demonstrate that many of the reports use nonspecific indicators to diagnose treponemal disease, do not provide adequate information about the methods used to date specimens, and do not include high-quality photographs of the lesions of interest. Thus, despite an increasing number of published reports of pre-Columbian treponemal infection, it appears that solid evidence supporting an Old World origin for the disease remains absent.
Link
A new comprehensive survey of the evidence, however, suggests that syphilis did come from the Americas due to Columbus' voyage, and that the evidence advanced for a pre-Columbian presence of the disease is not solid.
More:
“This is the first time that all 54 of these cases have been evaluated systematically,” says George Armelagos, an anthropologist at Emory University and co-author of the appraisal. “The evidence keeps accumulating that a progenitor of syphilis came from the New World with Columbus’ crew and rapidly evolved into the venereal disease that remains with us today.”
The appraisal was led by two of Armelagos’ former graduate students at Emory: Molly Zuckerman, who is now an assistant professor at Mississippi State University, and Kristin Harper, currently a post-doctoral fellow at Columbia University. Additional authors include Emory anthropologist John Kingston and Megan Harper from the University of Missouri.
“Syphilis has been around for 500 years,” Zuckerman says. “People started debating where it came from shortly afterwards, and they haven’t stopped since. It was one of the first global diseases, and understanding where it came from and how it spread may help us combat diseases today.”
Yrbk Phys Anthropol 54:99–133, 2011.
The origin and antiquity of syphilis revisited: An Appraisal of Old World pre-Columbian evidence for treponemal infection
Kristin N. Harper et al.
For nearly 500 years, scholars have argued about the origin and antiquity of syphilis. Did Columbus bring the disease from the New World to the Old World? Or did syphilis exist in the Old World before 1493? Here, we evaluate all 54 published reports of pre-Columbian, Old World treponemal disease using a standardized, systematic approach. The certainty of diagnosis and dating of each case is considered, and novel information pertinent to the dating of these cases, including radiocarbon dates, is presented. Among the reports, we did not find a single case of Old World treponemal disease that has both a certain diagnosis and a secure pre-Columbian date. We also demonstrate that many of the reports use nonspecific indicators to diagnose treponemal disease, do not provide adequate information about the methods used to date specimens, and do not include high-quality photographs of the lesions of interest. Thus, despite an increasing number of published reports of pre-Columbian treponemal infection, it appears that solid evidence supporting an Old World origin for the disease remains absent.
Link
December 19, 2011
Neandertal admixture: why I remain skeptical
The announcement by 23andMe of a Neandertal admixture feature of their commercial test gives me an opportunity to revisit the question of Neandertal admixture in general. At the outset, let me state that I'm still on the fence on whether there has been such admixture in Eurasians. The evidence that has appeared since the publication of Green et al. (2010) provides arguments both in favor and against the Neandertal admixture hypothesis.
Let's begin by examining the case for Neandertal introgression into Eurasians. This case boils down to the fact that modern Eurasians are more similar to Neandertals than modern Africans are. If Neandertals were an irrelevant outgroup, this is an unexpected finding.
The above statement in bold is the fact. But, this fact does not admit to the single interpretation of Neandertal admixture in the ancestors of Eurasians.
At the very beginning, I suggested that this fact could be explained by archaic admixture in Africans. Both genetics and paleoanthropology has furnished evidence in favor of my idea. It is no longer tenable to propose that Eurasians are shifted towards Neandertals only because of Neandertal admixture: in fact some of the shift may be due to Africans being shifted away from Neandertals because of admixture with archaic African hominins. Any future work on the issue must take this possibility into account.
A different pitfall is in the direction of gene flow: whether Neandertals donated genes to the Eurasian gene pool or vice versa. Again, I have contended that it is more likely for a successful expanding species to donate to a contracting species, rather than opposite. However, Green et al. proposed an ingenuous argument against that direction of gene flow:
The main idea is the following:
- Yoruba Nigerians are closer to Eurasians than San are.
- If the Neandertal genome is Proto-Eurasian-admixed, then it should be shifted towards Yoruba relative to San
- It does not appear to be, hence, on balance, gene flow was from Neandertals to modern humans, rather than the opposite.
The idea is fleshed out in the supplement of the Green et al. paper. It exploits the fact that modern human populations are not equidistant to each other, to show that an archaic hominin that was admixed with a subset of modern humans (Eurasians) would not only be shifted towards that population, but would also appear closer to populations close to Eurasians (=Yoruba), rather to those who are not (=San).
All this depends, of course, on the idea that the people who interbred with Neandertals were Proto-Eurasians, i.e., a subset of Africans who left the continent and went on to become modern Eurasians.
This idea is not as secure as it formerly appeared to be. The recognition of the real possibility of Out-of-Arabia means that the people who admixed with Neandertals may not have been Proto-Eurasians, but, rather undifferentiated Proto-Humans. In other words, they were not necessarily closer to modern Eurasians than to modern Africans, but, rather, common ancestors to both.
In conclusion:
I'm willing to bet that once scientists turn their attentions to full genomes, they will have substantial and indisputable evidence for genetic divergence between stretches of human DNA that simply too deep to be explained in a conventional Out-of-Africa timeframe.
If there was substantial archaic admixture in Africa c. 35ka, according to Hammer et al.'s estimate, and coinciding with the (intrusive?) appearance of Upper Paleolithic modern humans such as Hofmeyr, then full genome sequencing will provide the smoking gun evidence for it. Such an event would simultaneously solve many mysteries about the African population, such as its apparent higher effective population size, greater allele diversity, and recombination rate.
It may very well be that some level of Neandertal admixture will remain part of the story. We shall see.
Let's begin by examining the case for Neandertal introgression into Eurasians. This case boils down to the fact that modern Eurasians are more similar to Neandertals than modern Africans are. If Neandertals were an irrelevant outgroup, this is an unexpected finding.
The above statement in bold is the fact. But, this fact does not admit to the single interpretation of Neandertal admixture in the ancestors of Eurasians.
At the very beginning, I suggested that this fact could be explained by archaic admixture in Africans. Both genetics and paleoanthropology has furnished evidence in favor of my idea. It is no longer tenable to propose that Eurasians are shifted towards Neandertals only because of Neandertal admixture: in fact some of the shift may be due to Africans being shifted away from Neandertals because of admixture with archaic African hominins. Any future work on the issue must take this possibility into account.
A different pitfall is in the direction of gene flow: whether Neandertals donated genes to the Eurasian gene pool or vice versa. Again, I have contended that it is more likely for a successful expanding species to donate to a contracting species, rather than opposite. However, Green et al. proposed an ingenuous argument against that direction of gene flow:
The main idea is the following:
- Yoruba Nigerians are closer to Eurasians than San are.
- If the Neandertal genome is Proto-Eurasian-admixed, then it should be shifted towards Yoruba relative to San
- It does not appear to be, hence, on balance, gene flow was from Neandertals to modern humans, rather than the opposite.
The idea is fleshed out in the supplement of the Green et al. paper. It exploits the fact that modern human populations are not equidistant to each other, to show that an archaic hominin that was admixed with a subset of modern humans (Eurasians) would not only be shifted towards that population, but would also appear closer to populations close to Eurasians (=Yoruba), rather to those who are not (=San).
All this depends, of course, on the idea that the people who interbred with Neandertals were Proto-Eurasians, i.e., a subset of Africans who left the continent and went on to become modern Eurasians.
This idea is not as secure as it formerly appeared to be. The recognition of the real possibility of Out-of-Arabia means that the people who admixed with Neandertals may not have been Proto-Eurasians, but, rather undifferentiated Proto-Humans. In other words, they were not necessarily closer to modern Eurasians than to modern Africans, but, rather, common ancestors to both.
In conclusion:
- The inference of Neandertal admixture in modern Eurasians in terms of the D-statistic is proven to be a simplification that ignores archaic admixture in Africa
- The inference of Neandertal-to-modern admixture is based on the assumption that moderns admixing with Neandertals were already Eurasian-like, but the mounting evidence for a major human expansion Out-of-Arabia may mean that they were not.
I'm willing to bet that once scientists turn their attentions to full genomes, they will have substantial and indisputable evidence for genetic divergence between stretches of human DNA that simply too deep to be explained in a conventional Out-of-Africa timeframe.
If there was substantial archaic admixture in Africa c. 35ka, according to Hammer et al.'s estimate, and coinciding with the (intrusive?) appearance of Upper Paleolithic modern humans such as Hofmeyr, then full genome sequencing will provide the smoking gun evidence for it. Such an event would simultaneously solve many mysteries about the African population, such as its apparent higher effective population size, greater allele diversity, and recombination rate.
It may very well be that some level of Neandertal admixture will remain part of the story. We shall see.
December 18, 2011
Modern human vs. Neandertal brains
The long-term trend in human evolution has been towards larger brains. Neandertals, however, had somewhat larger brains than us. It turns out that modern humans surpassed Neandertals in the development of some areas of the brain.
Nature Communications 2, Article number: 588 doi:10.1038/ncomms1593
Evolution of the base of the brain in highly encephalized human species
Markus Bastir et al.
The increase of brain size relative to body size—encephalization—is intimately linked with human evolution. However, two genetically different evolutionary lineages, Neanderthals and modern humans, have produced similarly large-brained human species. Thus, understanding human brain evolution should include research into specific cerebral reorganization, possibly reflected by brain shape changes. Here we exploit developmental integration between the brain and its underlying skeletal base to test hypotheses about brain evolution in Homo. Three-dimensional geometric morphometric analyses of endobasicranial shape reveal previously undocumented details of evolutionary changes in Homo sapiens. Larger olfactory bulbs, relatively wider orbitofrontal cortex, relatively increased and forward projecting temporal lobe poles appear unique to modern humans. Such brain reorganization, beside physical consequences for overall skull shape, might have contributed to the evolution of H. sapiens' learning and social capacities, in which higher olfactory functions and its cognitive, neurological behavioral implications could have been hitherto underestimated factors.
Link
Nature Communications 2, Article number: 588 doi:10.1038/ncomms1593
Evolution of the base of the brain in highly encephalized human species
Markus Bastir et al.
The increase of brain size relative to body size—encephalization—is intimately linked with human evolution. However, two genetically different evolutionary lineages, Neanderthals and modern humans, have produced similarly large-brained human species. Thus, understanding human brain evolution should include research into specific cerebral reorganization, possibly reflected by brain shape changes. Here we exploit developmental integration between the brain and its underlying skeletal base to test hypotheses about brain evolution in Homo. Three-dimensional geometric morphometric analyses of endobasicranial shape reveal previously undocumented details of evolutionary changes in Homo sapiens. Larger olfactory bulbs, relatively wider orbitofrontal cortex, relatively increased and forward projecting temporal lobe poles appear unique to modern humans. Such brain reorganization, beside physical consequences for overall skull shape, might have contributed to the evolution of H. sapiens' learning and social capacities, in which higher olfactory functions and its cognitive, neurological behavioral implications could have been hitherto underestimated factors.
Link
December 16, 2011
First assessment of 1000 Genomes Iberian Spanish (IBS) sub-populations
As promised, I have taken the IBS sample from the latest available 1000 Genomes data and split it into sub-populations. There are at present 147 IBS individuals, and 108 of them have regional information about them:
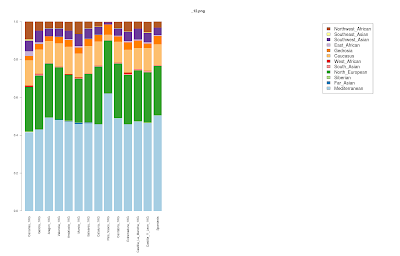
The one population that stands out in this set is that of the Basque Country (Pais_Vasco_1KG) which appears, like the HGDP French_Basque population to differ from its neighbors in having near zero of the "Caucasus" component.
I first speculated that some of the IBS sample were of Basque origin during the summer, and it seems that this was indeed the case.
The paucity of the "Caucasus" component in Dodecad, HGDP, and now 1000 Genomes Basques, together with the paucity of the "Caucasus"/"Gedrosia" components in Finns compared to northern Balto-Slavs and Scandinavians respectively is very suggestive, since these are the two major non-Indo-European speaking populations of Europe. (*)
In my opinion, these comparisons add weight to the growing body of evidence that the PIE Urheimat is to be sought in the territory of West Asia, as a secondary movement, about 8,000 years, ago of the broader series of expansions that began from this area 12,000 years ago.
(*) Hungarians are also non-Indo-European, but they seem to have received their language in historical times through a process of elite dominance.
- Canarias_1KG 2
- Galicia_1KG 8
- Aragon_1KG 6
- Valencia_1KG 12
- Andalucia_1KG 4
- Murcia_1KG 8
- Baleares_1KG 7
- Cataluna_1KG 9
- Pais_Vasco_1KG 8
- Cantabria_1KG 6
- Extremadura_1KG 8
- Castilla_La_Mancha_1KG 6
- Castilla_Y_Leon_1KG 12

The one population that stands out in this set is that of the Basque Country (Pais_Vasco_1KG) which appears, like the HGDP French_Basque population to differ from its neighbors in having near zero of the "Caucasus" component.
I first speculated that some of the IBS sample were of Basque origin during the summer, and it seems that this was indeed the case.
The paucity of the "Caucasus" component in Dodecad, HGDP, and now 1000 Genomes Basques, together with the paucity of the "Caucasus"/"Gedrosia" components in Finns compared to northern Balto-Slavs and Scandinavians respectively is very suggestive, since these are the two major non-Indo-European speaking populations of Europe. (*)
In my opinion, these comparisons add weight to the growing body of evidence that the PIE Urheimat is to be sought in the territory of West Asia, as a secondary movement, about 8,000 years, ago of the broader series of expansions that began from this area 12,000 years ago.
(*) Hungarians are also non-Indo-European, but they seem to have received their language in historical times through a process of elite dominance.
1000 Genomes at 2,100+ and counting
The latest working data on 1000 Genomes data include 2,123 individuals. I had already included some Khmer Vietnamese (KHV) from the previous working data for use with my K12a calculator. The list of populations in the datafile currently include:
GBR FIN CHS PUR CDX CLM IBS PEL KHV ACB CEU CHD YRI CHB JPT LWK ASW MXL TSI GIH MKK
I will probably take the time to extract anew the population data from the newest file, as well as split some (such as IBS) for which I have some more regional information. By my last count, I now have about ~10,600 individuals to work with (some are duplicates, e.g., between the HapMap and 1000 Genomes Project).
In other news, I see some 23/11/2011 data on Y-chromosome SNPs. I haven't worked on those myself, but I know that many hobbyists are interested in the Y-chromosome aspect of the project, so those might be useful.
Finally, there are slides from the ICHG seminar on the 1000 Genomes Project, which should be interesting reading.
Mega-study on Mexican admixture
There have been other studies on Mexican admixture patterns before, but this one breaks new ground, by determining the more specific origin of the three major components of the Mexican population. From the paper:
In addition to the HapMap CEU, who are mostly of Northern European ancestry, we used individuals recruited from Dublin, (Ireland), Warsaw (Poland), Rome (Italy) and Porto (Portugal) to provide references for different areas within Europe. The first two PCs provide good separation of these reference populations, and correspond roughly to North-South and West-East gradients (Figure 3A). Both the MEX1EUR and MEX2EUR virtual genomes are most closely related to intact genomes from Porto, which we interpret as a surrogate for populations from the Iberian Peninsula, [3], consistent with the historical record that the first European migrants to Mexico were Spaniards.The paper is also methodologically interesting:
Continental-level admixture proportions were estimated two ways: (1) a model-based clustering algorithm implemented in frappe [35], and (2) average locus-specific ancestries across all markers. Locus-specific ancestry was estimated with SABER+, an extension of a previously described approach, SABER, that uses a Markov-Hidden Markov Model [12]. SABER+ differs from SABER in implementation of a new algorithm, an Autoregressive Hidden Markov Model (ARHMM), in which haplotype structure within the ancestral populations is adaptively constructed using a binary decision tree based on as many as 15 markers, and which therefore does not require a priori knowledge of genome-wide ancestry proportions (Johnson et al., in preparation). In simulation studies, the ARHMM achieves accuracy comparable to HapMix [36] but is more flexible in modeling the three-way admixture in the Mexican population and does not require information about the recombination rate.PLoS Genet 7(12): e1002410. doi:10.1371/journal.pgen.1002410 Ancestral Components of Admixed Genomes in a Mexican Cohort Nicholas A. Johnson et al. For most of the world, human genome structure at a population level is shaped by interplay between ancient geographic isolation and more recent demographic shifts, factors that are captured by the concepts of biogeographic ancestry and admixture, respectively. The ancestry of non-admixed individuals can often be traced to a specific population in a precise region, but current approaches for studying admixed individuals generally yield coarse information in which genome ancestry proportions are identified according to continent of origin. Here we introduce a new analytic strategy for this problem that allows fine-grained characterization of admixed individuals with respect to both geographic and genomic coordinates. Ancestry segments from different continents, identified with a probabilistic model, are used to construct and study “virtual genomes” of admixed individuals. We apply this approach to a cohort of 492 parent–offspring trios from Mexico City. The relative contributions from the three continental-level ancestral populations—Africa, Europe, and America—vary substantially between individuals, and the distribution of haplotype block length suggests an admixing time of 10–15 generations. The European and Indigenous American virtual genomes of each Mexican individual can be traced to precise regions within each continent, and they reveal a gradient of Amerindian ancestry between indigenous people of southwestern Mexico and Mayans of the Yucatan Peninsula. This contrasts sharply with the African roots of African Americans, which have been characterized by a uniform mixing of multiple West African populations. We also use the virtual European and Indigenous American genomes to search for the signatures of selection in the ancestral populations, and we identify previously known targets of selection in other populations, as well as new candidate loci. The ability to infer precise ancestral components of admixed genomes will facilitate studies of disease-related phenotypes and will allow new insight into the adaptive and demographic history of indigenous people. Link
December 14, 2011
Clusters Galore analysis of West Eurasians
It's been a while since the last Clusters Galore analysis, so I've decided to use my recently assembled dataset and run such an analysis over the individuals who belonged to the Six main West Eurasian components.
Hence, at the beginning, I identified 945 individuals in my set who had more than 95% combined admixture proportions in the Six. Subsequently, I ran MDS on this set, keeping 50 dimensions.
One of the open issues in Clusters Galore analysis is how to choose how many MDS dimensions to retain. So far, I've applied a heuristic by choosing the number of MDS dimensions that maximizes the number of inferred clusters by MCLUST. However, when I actually inspect the MDS plots, it often turns out that meaningful information seems present at even higher number of MDS dimensions. As a result, I've decided to pick the number of dimensions in the following manner.
The main idea is that data points in uninformative MDS dimensions will appear as largely Gaussian noise. So, we can use a test of normality (I've chosen the Shapiro-Wilk test) to detect dimensions that appear not to be noise. Below is the p-value of this test for different MDS dimensions:
Up to 22 dimensions, there is a strong non-Gaussian signal (all p-values less than 0.001). Hence, I would use the first 22 dimensions in MCLUST analysis. With these dimensions, the number of inferred clusters was estimated as 35. So, this is something like a 6-fold increase in resolution over the Six components inferred by ADMIXTURE.
The cluster totals for the different populations can be seen in the spreadsheet.
Important Caveat: Some populations (e.g., Finnish_D, or Turkish_D) have a great number of individuals who do not meet the "95% in the Six" inclusion threshold. Hence, results are not representative for them, and simply indicate the cluster assignment of their subsets that do meet the threshold. You can check whether individuals have been removed from the original dataset by comparing sample sizes in the Clusters Galore spreadsheet with the K12a one.
Here are some observations on the 35 cluster. I will mention the modal population (or region) for each one:
- Ashkenazi
- Scandinavian
- French
- British Isles
- Armenian
- S Italian/Sicilian
- Kurd
- Greek
- Cypriot
- Balto-Slavic
- Hungarian
- Balkan
- Sephardic
- Spanish
- Iberian
- North Italian/Tuscan
- Morocco Jews (main)
- Saudis
- Georgian/Abkhazian
- Basque
- Bedouin
- Druze #1
- Druze #2
- Druze (main)
- Mozabite (main)
- Mozabite #1
- Orkney
- Sardinian
- Azerbaijan Jews
- Iran/Iraq Jews
- Lezgins
- Morocco Jews #1
- Samaritan
- Yemen Jews
- Abkhazian
Segmented HAPlotype Estimation and Imputation Tool (Delaneau et al. 2011)
The ShapeIT website seems to be very well-designed and informative.
Nature Methods (2011) doi:10.1038/nmeth.1785
A linear complexity phasing method for thousands of genomes
Olivier Delaneau et al.
Human-disease etiology can be better understood with phase information about diploid sequences. We present a method for estimating haplotypes, using genotype data from unrelated samples or small nuclear families, that leads to improved accuracy and speed compared to several widely used methods. The method, segmented haplotype estimation and imputation tool (SHAPEIT), scales linearly with the number of haplotypes used in each iteration and can be run efficiently on whole chromosomes.
Link
Nature Methods (2011) doi:10.1038/nmeth.1785
A linear complexity phasing method for thousands of genomes
Olivier Delaneau et al.
Human-disease etiology can be better understood with phase information about diploid sequences. We present a method for estimating haplotypes, using genotype data from unrelated samples or small nuclear families, that leads to improved accuracy and speed compared to several widely used methods. The method, segmented haplotype estimation and imputation tool (SHAPEIT), scales linearly with the number of haplotypes used in each iteration and can be run efficiently on whole chromosomes.
Link
December 13, 2011
Of Elephants and Men
The same team described dental remains from Qesem cave.
The press release:
It would be useful to get something more substantial than teeth to estimate the presence of a new hominin lineage as proposed by the authors. In their earlier work they noted some Neandertal traits in their teeth sample, although, on balance, they linked the Qesem pre-200ka teeth with the much later modern humans from the Levant. The 400ka date is (roughly) when geneticists tell us modern humans and Neandertals had already began to diverge genetically.The elephant, a huge package of food that is easy to hunt, disappeared from the Middle East 400,000 years ago -- an event that must have imposed considerable nutritional stress on Homo erectus. Working with Prof. Israel Hershkovitz of TAU's Sackler Faculty of Medicine, the researchers connected this evidence about diet with other cultural and anatomical clues and concluded that the new hominids recently discovered at Qesem Cave in Israel -- who had to be more agile and knowledgeable to satisfy their dietary needs with smaller and faster prey -- took over the Middle Eastern landscape and eventually replaced Homo erectus.The findings, which have been reported in the journal PLoS One, suggest that the disappearance of elephants 400,000 years ago was the reason that modern humans first appeared in the Middle East. In Africa, elephants disappeared from archaeological sites and Homo sapiens emerged much later -- only 200,000 years ago.
Neandertal-like traits have been noted in European hominins (such as Atapuerca) well in advance of this age. A priori, the idea that the common ancestor of modern humans and Neandertals lived in the Near East seems attractive to me, since that is the area where the demarcation line between the two species appears to have been. While the authors' theory about the disappearance of elephants and the emergence of the Acheulo-Yabrudian merits attention, I don't think the evidence for it is strong enough yet.
PLoS ONE 6(12): e28689. doi:10.1371/journal.pone.0028689
Man the Fat Hunter: The Demise of Homo erectus and the Emergence of a New Hominin Lineage in the Middle Pleistocene (ca. 400 kyr) Levant
Miki Ben-Dor et al.
The worldwide association of H. erectus with elephants is well documented and so is the preference of humans for fat as a source of energy. We show that rather than a matter of preference, H. erectus in the Levant was dependent on both elephants and fat for his survival. The disappearance of elephants from the Levant some 400 kyr ago coincides with the appearance of a new and innovative local cultural complex – the Levantine Acheulo-Yabrudian and, as is evident from teeth recently found in the Acheulo-Yabrudian 400-200 kyr site of Qesem Cave, the replacement of H. erectus by a new hominin. We employ a bio-energetic model to present a hypothesis that the disappearance of the elephants, which created a need to hunt an increased number of smaller and faster animals while maintaining an adequate fat content in the diet, was the evolutionary drive behind the emergence of the lighter, more agile, and cognitively capable hominins. Qesem Cave thus provides a rare opportunity to study the mechanisms that underlie the emergence of our post-erectus ancestors, the fat hunters.
Link
PLoS ONE 6(12): e28689. doi:10.1371/journal.pone.0028689
Man the Fat Hunter: The Demise of Homo erectus and the Emergence of a New Hominin Lineage in the Middle Pleistocene (ca. 400 kyr) Levant
Miki Ben-Dor et al.
The worldwide association of H. erectus with elephants is well documented and so is the preference of humans for fat as a source of energy. We show that rather than a matter of preference, H. erectus in the Levant was dependent on both elephants and fat for his survival. The disappearance of elephants from the Levant some 400 kyr ago coincides with the appearance of a new and innovative local cultural complex – the Levantine Acheulo-Yabrudian and, as is evident from teeth recently found in the Acheulo-Yabrudian 400-200 kyr site of Qesem Cave, the replacement of H. erectus by a new hominin. We employ a bio-energetic model to present a hypothesis that the disappearance of the elephants, which created a need to hunt an increased number of smaller and faster animals while maintaining an adequate fat content in the diet, was the evolutionary drive behind the emergence of the lighter, more agile, and cognitively capable hominins. Qesem Cave thus provides a rare opportunity to study the mechanisms that underlie the emergence of our post-erectus ancestors, the fat hunters.
Link
December 12, 2011
The womb of nations: how West Eurasians came to be
One of the most interesting observations about the genetic structure of the Old World, is the fact that distantly located populations are often more similar to each other than to their more immediate geographic neighbors.
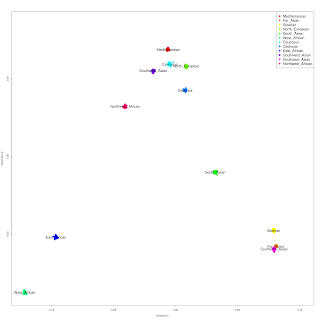



For example, in my recent K12a admixture experiment, the six components. I named Mediterranean, North_European, Caucasus, Gedrosia, Southwest_Asian, and Northwest_African have a maximum Fst between any two of them of 0.073 (between Gedrosia and Northwest_African), and a mimimum Fst between any of them and any of the others of 0.075 (between Gedrosia and South_Asian). Henceforth, I will call these six components simply "the Six."
It is remarkable that "Gedrosia", the component peaking in present-day Balochistan is about equidistant to the component peaking in Mozabite Berbers ("Northwest_African") and that peaking in South Indian Dravidian speakers ("South_Asian"). Going by geography alone, and even if we calculated distances "as the crow flies" and ignored all natural obstacles between Balochistan and the Sahara, we would have expected "Gedrosia" to be about 3 times more distant to "Northwest_African" than to "South_Asian".
Divergence between populations across the entire genome builds up mainly by two processes:
- Genetic drift
- Admixture
Moreover, if these populations migrate from their original homeland and absorb the indigenous inhabitants wherever they go, then they will diverge even more, depending on how much admixture they undergo, and how distantly related the aboriginal inhabitants are.

It is clear that admixture has played a role in the overall divergence of the Six. For example, the Northwest_African component is shifted (relative to the remaining five) towards the other African components; the Southwest_Asian is also thus shifted, but less noticeably. The Gedrosia component is shifted towards the South_Asian one; and the easternmost components, the North_European, and Gedrosia ones, are shifted towards the Asian components.
All these shifts are quite salient in the MDS plot, and there is ample evidence for the aboriginal populations being shifted in the expected direction in each region.
We can calculate the median Fst within the Six: it is 0.053, as well as the median Fst between members of the Six and all the rest: it is 0.13. The ratio of the two is ~40%. Of course, the various components have diverged from each other at different times: West and East Eurasians, for example, began diverging shortly after both diverged from Africans. Nonetheless, we can (conservatively) place the divergence time of the ancestors of the Six from East Asians, Ancestral South Indians, and Sub-Saharan Africans, at around 40,000 years ago, the time when the Upper Paleolithic (and modern man) makes its appearance all over the Old World.
Actual divergence times may be lower, and this is why 40k is a conservative estimate. The point of this back-of-a-napkin calculation is to give a rough estimate, rather than a precise date.
Assuming that Fst builds up roughly linearly with time due to drift (again a simplification), we can estimate that divergence within the Six dates to less than 16 thousand years ago. This may be an overestimate for two reasons:
- Divergence between Proto-West Eurasians and the rest of mankind may be less than 40k years
- Each of the Six have partially absorbed aboriginal inhabitants (e.g., Palaeo-Europeans, Ancestral South Indians, Pre-Berber North Africans, etc.) that spent most of the Paleolithic diverging from the common ancestors of the Six.
The womb of nations

The Neolithic of West Eurasia started, by most accounts, c. 12 thousand years ago. Its origin was in the area framed by the Armenian Plateau in the north, the Anatolian Plateau in the west, the Zagros Range in the east, and the lowlands of southern Mesopotamia and the Levant in the south. Intriguingly, the prehistoric site of Göbekli Tepe sits right at the center of this important area, in eastern Anatolia/northern Mesopotamia.
If there is a candidate for where the ur-population that became the modern Six lived, the early Neolithic of the Near East is surely it. This hypothesis makes the most sense chronologically, archaeologically, genetically, and geographically.
Migrants out of the core area would have spread their genes in all directions, becoming differentiated by a combination of drift, admixture, and the selection pressures they faced in different natural and cultural environments; some of them would acquire lighter pigmentation, others lactase persistence, malaria resistence, the ability to process the dry desert air or to survive the long winter nights of the arctic. These spreads were sometimes gradual, sometimes dramatic: they took place over thousands of years and from a multitude of secondary and tertiary staging points.
In Arabia, the migrants would have met aboriginal Arabians, similar to their next door-neighbors in East Africa, undergoing a subtle African shift (Southwest_Asians). In North Africa, they would have encountered denser populations during the favorable conditions of MIS 1, and by absorbing them they would became the Berbers (Northwest_Africans). Their migrations to the southeast brought them into the realm of Indian-leaning people, in the rich agricultural fields of the Mehrgarh and the now deserted oases of Bactria and Margiana. Across the Mediterranean and along the Atlantic facade of Europe, they would have encountered the Mesolithic populations of Europe, and through their blending became the early Neolithic inhabitants of the Mediterranean and Atlantic coasts of Europe (Mediterraneans). And, to the north, from either the Balkans, the Caucasus, or the trans-Caspian region, they would have met the last remaining Proto-Europeoid hunters of the continental zone, becoming the Northern Europeoids who once stretched all the way to the interior of Asia.

It is, perhaps, in the ancient land of the Colchi, protected by the Black and Caspian seas, and by tall mountains on the remaining sides, that something resembling the ur-population survived. The great linguistic diversity of the Caucasian peoples, the central position of the Caucasus component among the Six (see MDS plot), and the fact that theirs was a remote region, ignored by the empires of the Near East to the south, and the frequent travellers of the Eurasiatic steppe to the north, may all indicate the plausibility of this assertion. Through a peculiar coincidence, old Blumenbach may have been onto something, although for reasons he could scarcely have imagined.
We don't have to suppose that a single process drove the dispersal of genes from the core area of the Near East. Agriculture was definitely an early facilitator of dispersal, but a variety of technological developments may have been instrumental in further spurts of dispersal: the invention of pottery, metalworking, pastoralism, sea navigation, the engines of war, all the way to the recent past, when ocean-going vessels from Western Europe sailed to the New World, Arab merchants and warriors spread their new religion to the continent of Africa, and Russian hunters and farmers began the conquest of Siberia.
There are hints that scientists are beginning to realize that this story may describe what happened during prehistory. The next few years will help resolve this grand puzzle of "how West Eurasians came to be."
Subscribe to:
Posts (Atom)